Genetic mosaicism, the presence of genetically distinct cell populations within an individual, arises due to somatic mutations occurring during embryogenesis and throughout life. This phenomenon is critical in both normal development and disease pathogenesis, particularly in the aging brain. Understanding mosaicism requires precise genomic analysis, which has been significantly advanced by single-cell whole-genome sequencing (WGS). However, accurate detection of somatic mutations at the single-cell level has been hindered by technical limitations in whole-genome amplification (WGA). Recent advances, including primary template-directed amplification (PTA), have greatly improved the fidelity of single-cell genomics.
Origins of Genetic Mosaicism
Mosaicism occurs when mutations arise in a subset of cells during early embryogenesis or later in life due to environmental stressors, replication errors, or DNA repair deficiencies. These mutations can be classified into several types:
- Point mutations: Single nucleotide changes that may impact gene function.
- Copy number variations (CNVs): Duplications or deletions of large genomic segments.
- Structural variants: Large-scale genomic rearrangements such as translocations and inversions.
- Mobile element insertions: Retrotransposon activity leading to new DNA insertions.
Early mutations in embryogenesis can lead to tissue-specific mosaicism, which has implications for developmental disorders and cancer predisposition. In the aging brain, accumulated somatic mutations contribute to neurodegenerative diseases and cognitive decline (Miller et al., 2021). Due to their low allele frequencies, detecting these mutations requires ultra-sensitive genomic techniques.
The Necessity of Single-Cell Whole-Genome Sequencing
Traditional bulk sequencing averages genetic information across many cells, obscuring rare somatic mutations. Single-cell WGS allows for precisely identifying these mutations in individual cells, which is crucial for studying genetic heterogeneity in tissues like the brain, where neurons exhibit high mutation burdens with age (Lodato et al., 2018).
To perform single-cell WGS, whole-genome amplification (WGA) is necessary to amplify the minute DNA content of a single cell (∼6 pg). However, conventional WGA methods introduce amplification bias and errors, limiting their effectiveness in detecting somatic mutations.
Technical Limitations of Whole-Genome Amplification
Early WGA methods suffered from uneven genome coverage and artifacts, complicating mutation detection. The three primary WGA techniques used for single-cell WGS are:
- PicoPlex (Takara Bio)
- Uses preamplification followed by PCR amplification.
- Strengths: Low-input DNA requirement and streamlined workflow.
- Weaknesses: High allelic dropout and significant amplification bias (Takara Bio).
- REPLI-g (Qiagen)
- Based on multiple displacement amplification (MDA) using phi29 polymerase.
- Strengths: High coverage uniformity and minimal sequence bias.
- Weaknesses: Prone to chimeric artifacts and poor amplification of GC-rich regions (Qiagen; Gonzalez-Pena et al., 2021).
- Primary Template-Directed Amplification (PTA) (BioSkryb)
- Selectively amplifies original DNA templates rather than copied amplicons.
- Strengths: Significantly reduces errors and amplification bias, leading to higher accuracy in single-cell mutation detection (BioSkryb Technologies, 2024).
- Weaknesses: Slightly more expensive than REPLI-g.
Advances in Detecting Mosaicism with PTA
PTA has revolutionized single-cell WGS by maintaining high genome coverage while minimizing amplification errors. Compared to traditional WGA methods, PTA provides:
- Higher sensitivity for detecting rare somatic mutations.
- Improved uniformity across the genome, reducing false positives.
- Better representation of GC-rich and repetitive regions.
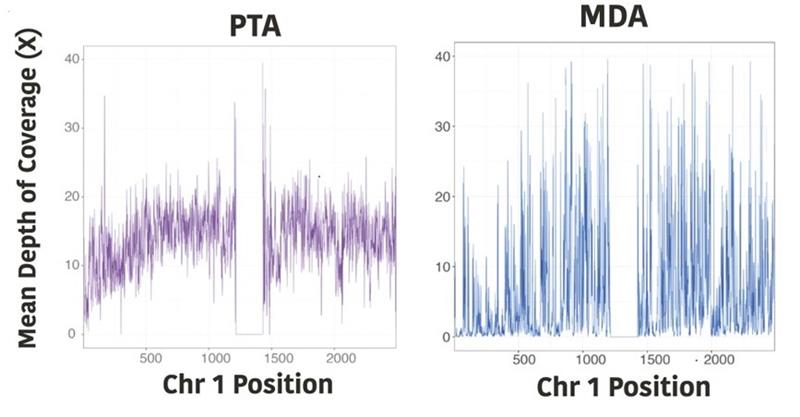
Fig 1 (from Gonzalez-Pena PNAS 2021)
In a study by Gonzalez-Pena et al., the authors show that PTA significantly improved uniformity of coverage compared to MDA (Figure 1). They also demonstrate that PTA approaches the two bulk (unamplified) samples at every coverage depth, which is a significant improvement over traditional MDA, which maxes out at 75% of the genome.
These advancements have profound implications for studying neurodevelopmental disorders, cancer evolution, and aging-related somatic mutation accumulation (Bae et al., 2018).
Conclusion
Genetic mosaicism is critical in human development and aging, necessitating accurate detection methods. Single-cell WGS has emerged as a powerful tool, but its success depends on reliable whole-genome amplification. While PicoPlex and REPLI-g have contributed to early single-cell studies, PTA has set a new standard by significantly improving accuracy. As sequencing technologies evolve, these advancements will enhance our understanding of somatic mutations in health and disease. PTA kits can be purchased from BioSkryb Genomics or run using a PTA service provider, like MiRXES. MiRXES is one of the few service providers certified to run the ResolveOME Whole Genome and Transcriptome Amplification Kit. The ResolveOME Whole Genome and Transcriptome Amplification Kit combines whole-genome and full-transcript transcriptome sequencing from the same cell. MiRXES also offers spatial transcriptomic services using Stereo-seq to combine spatial information with gene expression data.
References
- Miller MB, Reed HC, Walsh CA. Brain Somatic Mutation in Aging and Alzheimer’s Disease. Annual Review of Genomics Human Genetics. 2021 Aug 31;22:239-256.
- Lodato, M.A., et al. (2018). “Aging and neurodegeneration are associated with increased mutations in single human neurons.” Science, 359(6375), 555-559.
- Gonzalez-Pena, V., et al. (2021). “Accurate genomic variant detection in single cells with primary template-directed amplification.” PNAS, Jun 15;118(24).
- Bae, T., et al. (2018). “Different mutational rates and mechanisms in human cells at different developmental stages.” Science, 359(6375):550-555..
- BioSkryb Technologies. (2024). “Primary template-directed amplification (PTA).” Retrieved from [https://www.bioskryb.com/technology/]