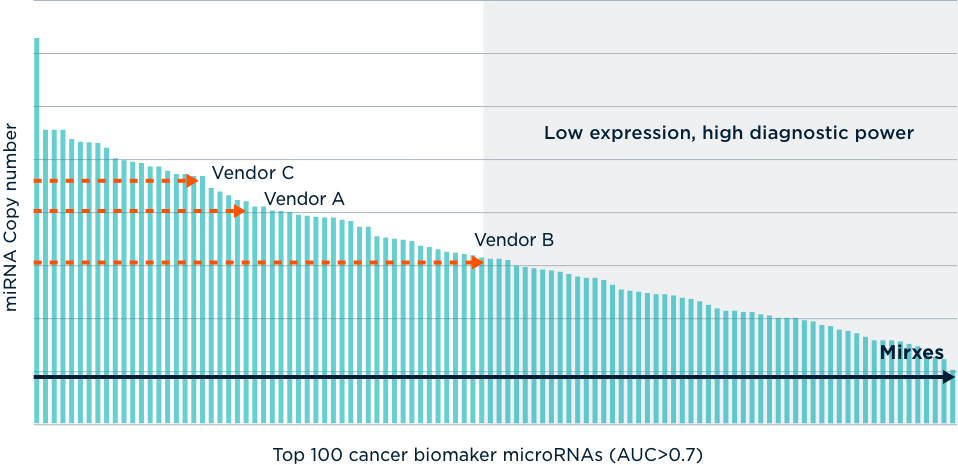
MicroRNAs (miRNAs), the smallest genetic material known, offer tremendous promise as biomarkers for early detection of diseases—from cancer and cardiovascular disease to metabolic, neurodegenerative, and infectious diseases. Mirxes offers advanced miRNA profiling capabilities to power your discovery and translational research.
While miRNAs play an essential role in regulating gene expression, they are difficult to detect accurately due to their small size. Our class-leading ID3EAL™ RT-qPCR technology overcomes this challenge, accurately detecting and quantifying miRNAs. Mirxes has also assembled the world’s largest clinical mRNA database, providing valuable insight for research.