DNA Nanoball Sequencing DNBSEQ™
DNA nanoball sequencing (DNBSEQ™) is a cutting-edge method for sequencing DNA that has gained popularity in recent years as an alternative to the bridge amplification method used by Illumina instruments. DNA nanoball sequencing uses a combination of nanotechnology, polymerase amplification, and fluorescent imaging to sequence DNA with high accuracy and throughput. One critical component of nanoball sequencing is the phi 29* polymerase, which plays a vital role in amplifying DNA fragments.
Like other members of the B family of DNA polymerases, Phi 29 can amplify DNA fragments with high fidelity and processivity. It can synthesize up to 70,000 bases per binding event and has robust strand displacement activity with a 3′→ 5′ proofreading exonuclease function. Phi 29 polymerase amplifies DNA fragments through rolling circle amplification (RCA). In RCA, a circular DNA library serves as a template to synthesize a single-stranded circular DNA molecule. This circular DNA molecule is then used by phi 29 polymerase to synthesize multiple copies of a complementary DNA strand. This process results in a long, branched DNA molecule called a nanoball. This nanoball contains thousands of copies of the same DNA fragment. This is in contrast to Illumina sequencing, which uses a lower fidelity polymerase and amplifies its template using bridge amplification. Both Illumina and DNA nanoball sequencing utilize the SBS method. SBS uses fluorescent imaging, where each base is detected as it is incorporated during DNA synthesis. Besides its high throughput and accuracy, another advantage of nanoball sequencing is its use of a chip containing evenly-spaced oligos. This approach is different from other sequencing technologies that use a randomly patterned flow cell (Fig 1).
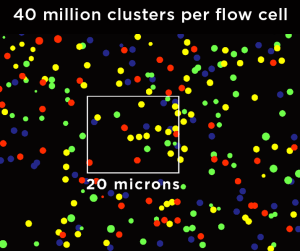
Technologies that use randomly patterned flow cells, like Illumina, tend to have areas on the flow cell with overcrowded clusters (over-clustered). For example, after the library solution is added, the library gets bound to the front portion of the flow cell. Then, the library concentration decreases as it flows to the back of the flow cell. In other situations, if the flow cell is overloaded (the picomolar concentration is too high), over-clustering occurs throughout the flow cell. Overcrowding can lead to multiple clusters being mistaken as a single cluster. This often results in noisy signals and drops in Q30 scores.
In conclusion, nanoball sequencing is an innovative technique for sequencing DNA that utilizes phi 29 polymerase to amplify DNA fragments through rolling circle amplification. Using a chip with evenly spaced oligos offers several advantages over a randomly patterned flow cell, including improved accuracy and coverage of the sequencing data. As sequencing technologies continue to evolve, nanoball sequencing is poised to become a powerful tool for genomics research and clinical applications.
*Phi 29 is not used by Illumina’s sequencing by the synthesis (SBS) method.