Stereo-seq Stands Out Among Spatial Transcriptomics Platforms
A recently published paper titled “An invasive zone in human liver cancer identified by Stereo-seq promotes hepatocyte-tumor cell crosstalk, local immunosuppression and tumor progression” by Wu et. al. shows how liver tumor cells interact with the surrounding microenvironment during cancer progression(1). The study, available in the journal Cancer Gene Therapy, utilizes a recently developed high resolution spatial transcriptomics platform, Stereo-seq, and illustrates the molecular dialogue occurring between cancerous hepatocytes and their neighboring healthy counterparts.
This new study demonstrates the power of Stereo-seq stands out among spatial transcriptomics platforms and its unique ability to provide sub-cellular high-resolution spatial gene expression profiles while retaining the spatial context of cells within the tissue. Other methods, like single-cell RNA sequencing (scRNA-seq), offer valuable insights into cellular behavior but lack the ability to capture this context. Conversely, techniques such as FISH offer spatial context but are limited in terms of the number of genes that can be probed simultaneously. Stereo-seq bridges this gap by fusing these approaches, thus offering an unparalleled level of information regarding the spatial distribution of gene expression across a wide spectrum of genes. In contrast, Stereo-seq allows researchers to not only detect individual RNA transcripts but also to pinpoint their spatial distribution within tissue sections.
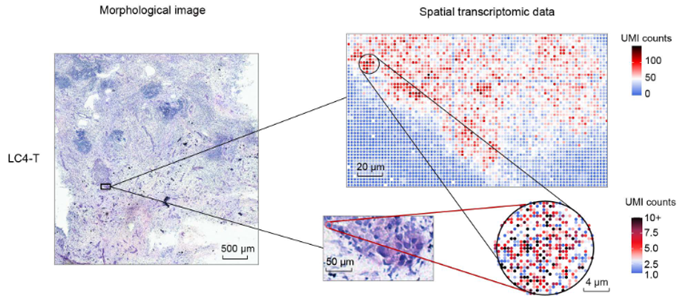
In this paper, the authors use Stereo-seq to study liver cancer. Liver cancers, including hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (ICC), are one of the most prevalent and challenging malignancies worldwide. They often exhibit aggressive behavior with limited treatment options. Traditional studies have predominantly focused on deciphering genetic mutations within tumor cells themselves, inadvertently sidelining the dynamic interplay between tumor and normal cells within the tumor microenvironment (TME). In this study, the authors generated Stereo-seq datasets from 21 patients who had been diagnosed with liver cancer (HCC, n = 6; ICC, n = 15). From each of these patient’s tumors multiple tumor regions were analyzed. This included tumor tissues (T, n=12), tumor margin areas (M, n=21), paratumor tissues (P, n=10), and normal or metastatic lymph nodes (LN, n=10) from 53 sub-samples. Their attention was focused on the margin areas (M) of the tumor as this is where the cancer cells are dividing and expanding into the paratumor region containing hepatocytes. Using Stereo-seq, they mapped transcripts back to the original tissues which were stained using hematoxylin and eosin (H&E). The hematoxylin stains cell nuclei and eosin stains cytoplasm. Once the sequences were mapped back to the tissue, they were binned into pseudo-spots, each representing one cell. Each cell, or pseudo-spot, contained an average of 589–4642 mRNA molecules and 366–1897 genes. For this report, their attention was focused on the margin areas (M) of the tumor where the cancer cells are dividing and expanding into the hepatocytes containing paratumor region.
In addition to Stereo-seq data, 3 of the 21 patients whose tumors were analyzed using Stereo-seq, also had scRNA seq data generated. . The scRNA seq data was used to independently verify cell types. Using Seurat(2), they were able to cluster gene expression profiles from 62,155 cells into 11 main cell types. The cell types they found were T cells (CD3D), natural killer cells (NKs) (KLRF1), B cells (MS4A1), plasma cells (MZB1), macrophages (CD163/CD14), dendritic cells (DCs) (CD1C), cholangiocytes or malignant cells (KRT19/EPCAM), hepatocytes (ALB), endothelial cells (CDH5/ENG), and fibroblasts (ACTA2). By combining Stereo-seq, scRNA-seq, and immunofluorescence data, they found that CXCL6 was being upregulated and secreted by invasive tumor cells. This ligand, activates the JAK-STAT3 pathway in nearby hepatocytes through the CXC Motif Chemokine Receptor 2 (CXCR2). This is consistent with previous reports that showed CXCL6 activates the JAK-STAT3 pathway (3,4). Activation of the JAK-STAT3 pathway was show to lead to the overexpression of SAAs in these hepatocytes using Single-Cell Regulatory Network Inference and Clustering (SCENIC) analysis(5) . Multiplexed immunofluorescence (IF) staining was used to confirm STAT3 expression levels in SAA+ hepatocytes. IF staining was also used to confirm the location of CXCL6+ tumor cells relative to SAA+ hepatocytes in the invasive zone.
Collectively, their results underscore the essential role of local hepatocyte-tumor cell crosstalk in fostering tumor progression. The study’s findings also have broader implications for cancer therapy. The observed local immunosuppression in the invasive zone provides a potential explanation for the immune system’s limited success in targeting liver tumors. By characterizing the complex interplay between tumor cells, hepatocytes, and immune cells, this research sets the stage for designing more effective therapeutic strategies that tackle both tumor cells and their supportive microenvironment. The findings in this paper not only shed light on previously unexplored aspects of liver cancer progression but also pave the way for novel therapeutic avenues. As the field of cancer research continues to evolve, Stereo-seq stands as a testament to the power of innovation in unraveling the complexities of human health and disease.
Keywords: Spatial Transcriptomics, RNA-seq, scRNA-seq, single cell, Next Generation Sequencing (NGS), Liver Cancer, hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma (ICC), Tumor Microenvironment (TME), JAK-STAT3 signaling pathway, hepatocytes, hematoxylin, eosin, mRNA
-
- Wu L, Yan J, Bai Y, Chen F, Zou X, Xu J, et al. An invasive zone in human liver cancer identified by Stereo-seq promotes hepatocyte-tumor cell crosstalk, local immunosuppression and tumor progression. Cell Res. 2023 Aug;33(8):585–603.
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015 May;33(5):495–502.
- Zheng S, Shen T, Liu Q, Liu T, Tuerxun A, Zhang Q, et al. CXCL6 fuels the growth and metastases of esophageal squamous cell carcinoma cells both in vitro and in vivo through upregulation of PD-L1 via activation of STAT3 pathway. J Cell Physiol. 2021 Jul;236(7):5373–86.
- Sun MY, Wang SJ, Li XQ, Shen YL, Lu JR, Tian XH, et al. CXCL6 Promotes Renal Interstitial Fibrosis in Diabetic Nephropathy by Activating JAK/STAT3 Signaling Pathway. Front Pharmacol. 2019;10:224.
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017 Nov;14(11):1083–6.